Publications
Publications by categories in reversed chronological order.
2025
-
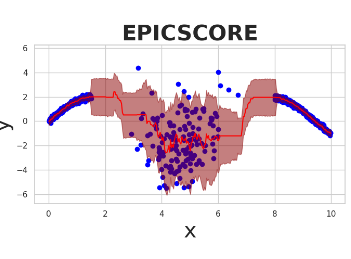 Epistemic Uncertainty in Conformal Scores: A Unified ApproachLuben Miguel Cruz Cabezas, Vagner Silva Santos, Thiago Ramos, and 1 more authorIn Proceedings of the Forty-first Conference on Uncertainty in Artificial Intelligence, 21–25 jul 2025
Epistemic Uncertainty in Conformal Scores: A Unified ApproachLuben Miguel Cruz Cabezas, Vagner Silva Santos, Thiago Ramos, and 1 more authorIn Proceedings of the Forty-first Conference on Uncertainty in Artificial Intelligence, 21–25 jul 2025Conformal prediction methods create prediction bands with distribution-free guarantees but do not explicitly capture epistemic uncertainty, which can lead to overconfident predictions in data-sparse regions. Although recent conformal scores have been developed to address this limitation, they are typically designed for specific tasks, such as regression or quantile regression. Moreover, they rely on particular modeling choices for epistemic uncertainty, restricting their applicability. We introduce EPICSCORE, a model-agnostic approach that enhances any conformal score by explicitly integrating epistemic uncertainty. Leveraging Bayesian techniques such as Gaussian Processes, Monte Carlo Dropout, or Bayesian Additive Regression Trees, EPICSCORE adaptively expands predictive intervals in regions with limited data while maintaining compact intervals where data is abundant. As with any conformal method, it preserves finite-sample marginal coverage. Additionally, it also achieves asymptotic conditional coverage. Experiments demonstrate its good performance compared to existing methods. Designed for compatibility with any Bayesian model, but equipped with distribution-free guarantees, EPICSCORE provides a general-purpose framework for uncertainty quantification in prediction problems.
@inproceedings{cabezas2025epistemicuncertaintyconformalscores, title = {Epistemic Uncertainty in Conformal Scores: A Unified Approach}, author = {Cruz Cabezas, Luben Miguel and Silva Santos, Vagner and Ramos, Thiago and Izbicki, Rafael}, booktitle = {Proceedings of the Forty-first Conference on Uncertainty in Artificial Intelligence}, pages = {443--470}, year = {2025}, editor = {Chiappa, Silvia and Magliacane, Sara}, volume = {286}, series = {Proceedings of Machine Learning Research}, month = {21--25 Jul}, publisher = {PMLR}, url = {https://proceedings.mlr.press/v286/cruz-cabezas25a.html} } -
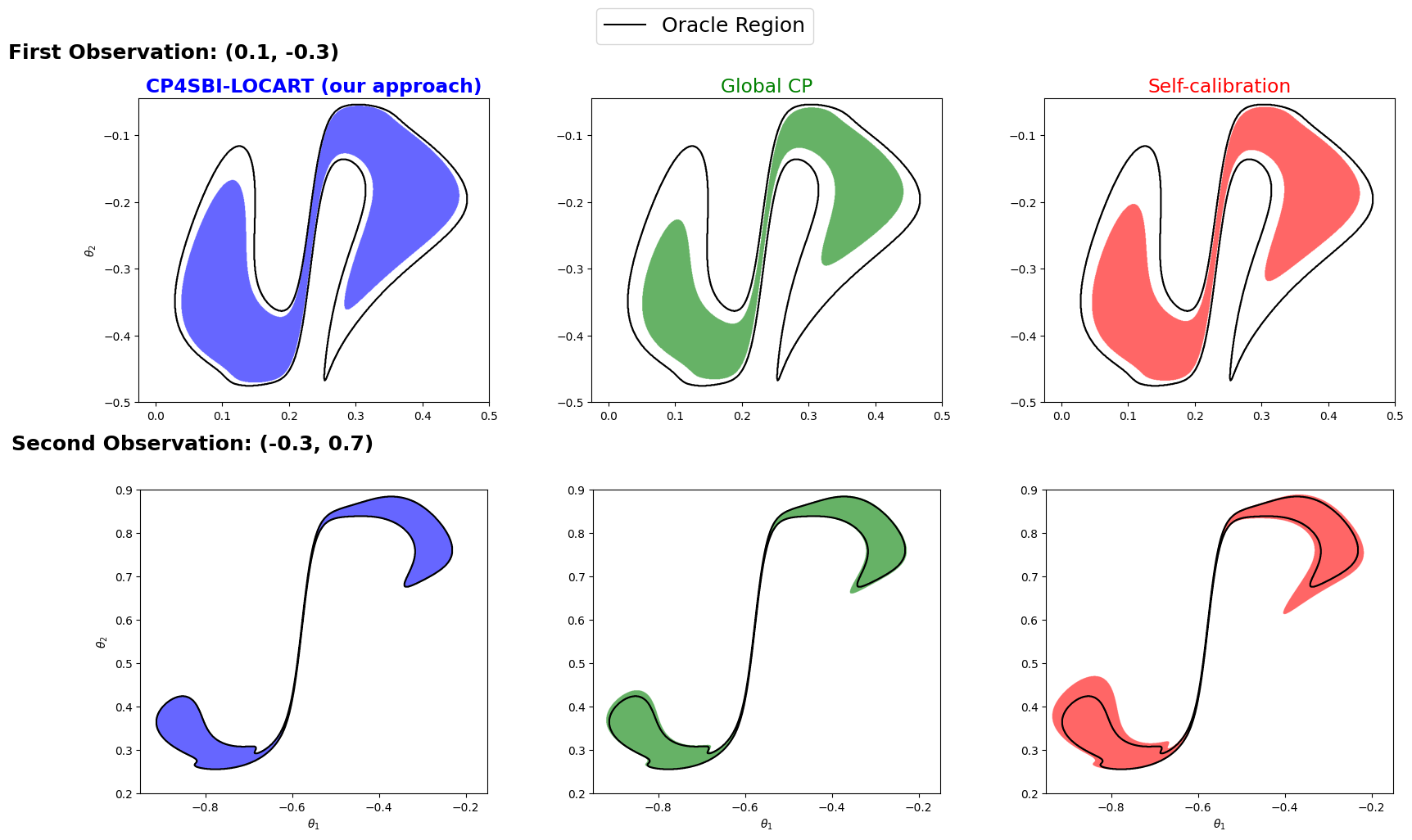 CP4SBI: Local Conformal Calibration of Credible Sets in Simulation-Based InferenceLuben M. C. Cabezas, Vagner S. Santos, Thiago R. Ramos, and 2 more authors2025
CP4SBI: Local Conformal Calibration of Credible Sets in Simulation-Based InferenceLuben M. C. Cabezas, Vagner S. Santos, Thiago R. Ramos, and 2 more authors2025Current experimental scientists have been increasingly relying on simulationbased inference (SBI) to invert complex non-linear models with intractable likelihoods. However, posterior approximations obtained with SBI are often miscalibrated, causing credible regions to undercover true parameters. We develop CP4SBI, a model-agnostic conformal calibration framework that constructs credible sets with local Bayesian coverage. Our two proposed variants, namely local calibration via regression trees and CDF-based calibration, enable finite-sample local coverage guarantees for any scoring function, including HPD, symmetric, and quantile-based regions. Experiments on widely used SBI benchmarks demonstrate that our approach improves the quality of uncertainty quantification for neural posterior estimators using both normalizing flows and score-diffusion modeling.
@article{cabezas2025cp4sbi, title = {CP4SBI: Local Conformal Calibration of Credible Sets in Simulation-Based Inference}, author = {Cabezas, Luben M. C. and Santos, Vagner S. and Ramos, Thiago R. and Rodrigues, Pedro L. C. and Izbicki, Rafael}, month = {}, year = {2025}, primaryclass = {stat.ML}, archiveprefix = {arXiv}, eprint = {2508.17077}, } -
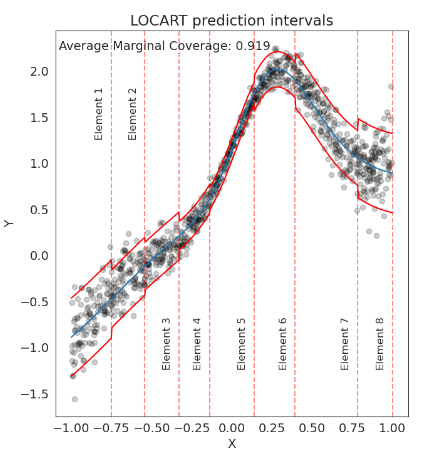 Regression trees for fast and adaptive prediction intervalsLuben MC Cabezas, Mateus P Otto, Rafael Izbicki, and 1 more authorInformation Sciences, 2025
Regression trees for fast and adaptive prediction intervalsLuben MC Cabezas, Mateus P Otto, Rafael Izbicki, and 1 more authorInformation Sciences, 2025In predictive modeling, quantifying prediction uncertainty is crucial for reliable decision-making. Traditional conformal inference methods provide marginally valid predictive regions but often produce non-adaptive intervals when naively applied to regression, potentially biasing applications. Recent advances using quantile regressors or conditional density estimators improve adaptability but are typically tied to specific prediction models, limiting their ability to quantify uncertainty around arbitrary models. Similarly, methods based on partitioning the feature space adopt sub-optimal strategies, failing to consistently measure predictive uncertainty across the feature space, especially in adversarial examples. This paper introduces a model-agnostic family of methods to calibrate prediction intervals for regression with local coverage guarantees. By leveraging regression trees and Random Forests, our approach constructs data-adaptive partitions of the feature space to approximate conditional coverage, enhancing the accuracy and scalability of prediction intervals. Our methods outperform established benchmarks on simulated and real-world datasets. They are implemented in the Python package clover, which integrates seamlessly with the scikit-learn interface for practical application.
@article{cabezas2025regression, title = {Regression trees for fast and adaptive prediction intervals}, author = {Cabezas, Luben MC and Otto, Mateus P and Izbicki, Rafael and Stern, Rafael B}, journal = {Information Sciences}, volume = {686}, pages = {121369}, year = {2025}, month = {}, publisher = {Elsevier}, } -
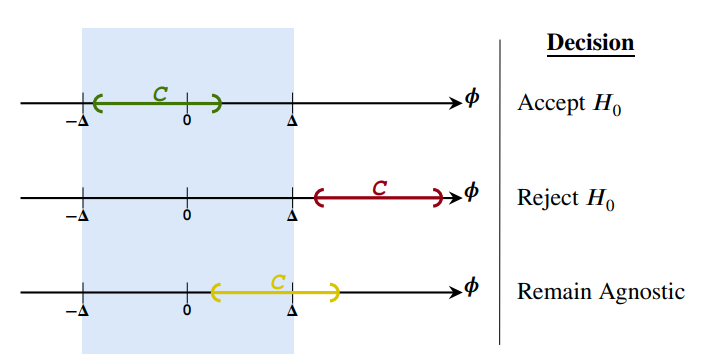 REACT to NHST: Sensible conclusions for meaningful hypothesesRafael Izbicki, Luben M. C. Cabezas, Fernando A. B. Colugnatti, and 3 more authorsThe Quantitative Methods for Psychology, 2025
REACT to NHST: Sensible conclusions for meaningful hypothesesRafael Izbicki, Luben M. C. Cabezas, Fernando A. B. Colugnatti, and 3 more authorsThe Quantitative Methods for Psychology, 2025While Null Hypothesis Significance Testing (NHST) remains a widely used statistical tool, it suffers from several shortcomings in its common usage, such as conflating statistical and practical significance, the formulation of inappropriate null hypotheses, and the inability to distinguish between accepting the null hypothesis and failing to reject it. Recent efforts have focused on developing alternatives that address these issues. Despite these efforts, conventional NHST remains dominant in scientific research due to its procedural simplicity and mistakenly presumed ease of interpretation. Our work presents an intuitive alternative to conventional NHST designed to bridge the gap between the expectations of researchers and the actual outcomes of hypothesis tests: REACT. REACT not only tackles shortcomings of conventional NHST but also offers additional advantages over existing alternatives. For instance, REACT applies to multiparametric hypotheses and does not require stringent significance-level corrections when conducting multiple tests. We illustrate the practical utility of REACT through real-world data examples.
2024
-
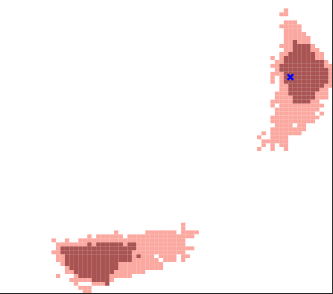 Distribution-Free Calibration of Statistical Confidence SetsLuben M. C. Cabezas, Guilherme P. Soares, Thiago R. Ramos, and 2 more authors2024
Distribution-Free Calibration of Statistical Confidence SetsLuben M. C. Cabezas, Guilherme P. Soares, Thiago R. Ramos, and 2 more authors2024Constructing valid confidence sets is a crucial task in statistical inference, yet traditional methods often face challenges when dealing with complex models or limited observed sample sizes. These challenges are frequently encountered in modern applications, such as Likelihood-Free Inference (LFI). In these settings, confidence sets may fail to maintain a confidence level close to the nominal value. In this paper, we introduce two novel methods, TRUST and TRUST++, for calibrating confidence sets to achieve distribution-free conditional coverage. These methods rely entirely on simulated data from the statistical model to perform calibration. Leveraging insights from conformal prediction techniques adapted to the statistical inference context, our methods ensure both finite-sample local coverage and asymptotic conditional coverage as the number of simulations increases, even if n is small. They effectively handle nuisance parameters and provide computationally efficient uncertainty quantification for the estimated confidence sets. This allows users to assess whether additional simulations are necessary for robust inference. Through theoretical analysis and experiments on models with both tractable and intractable likelihoods, we demonstrate that our methods outperform existing approaches, particularly in small-sample regimes. This work bridges the gap between conformal prediction and statistical inference, offering practical tools for constructing valid confidence sets in complex models.
@misc{cabezas2024distributionfreecalibrationstatisticalconfidence, title = {Distribution-Free Calibration of Statistical Confidence Sets}, author = {Cabezas, Luben M. C. and Soares, Guilherme P. and Ramos, Thiago R. and Stern, Rafael B. and Izbicki, Rafael}, year = {2024}, eprint = {2411.19368}, archiveprefix = {arXiv}, primaryclass = {stat.ME}, }
2023
-
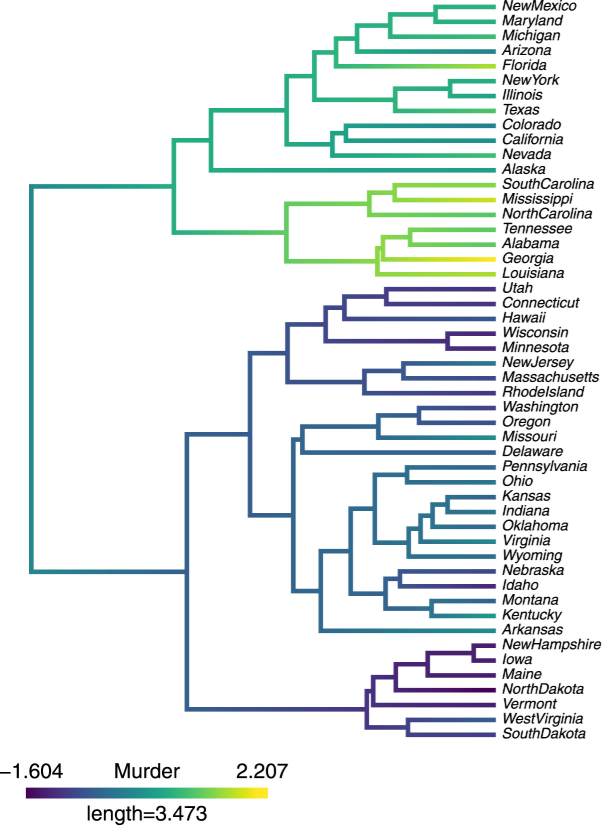 Hierarchical clustering: Visualization, feature importance and model selectionLuben MC Cabezas, Rafael Izbicki, and Rafael B SternApplied Soft Computing, 2023
Hierarchical clustering: Visualization, feature importance and model selectionLuben MC Cabezas, Rafael Izbicki, and Rafael B SternApplied Soft Computing, 2023We propose methods for the analysis of hierarchical clustering that fully use the multi-resolution structure provided by a dendrogram. Specifically, we propose a loss for choosing between clustering methods, a feature importance score and a graphical tool for visualizing the segmentation of features in a dendrogram. Current approaches to these tasks lead to loss of information since they require the user to generate a single partition of the instances by cutting the dendrogram at a specified level. Our proposed methods, instead, use the full structure of the dendrogram. The key insight behind the proposed methods is to view a dendrogram as a phylogeny. This analogy permits the assignment of a feature value to each internal node of a tree through an evolutionary model. Real and simulated datasets provide evidence that our proposed framework has desirable outcomes and gives more insights than state-of-art approaches. We provide an R package that implements our methods.
@article{cabezas2023hierarchical, title = {Hierarchical clustering: Visualization, feature importance and model selection}, author = {Cabezas, Luben MC and Izbicki, Rafael and Stern, Rafael B}, journal = {Applied Soft Computing}, volume = {141}, pages = {110303}, year = {2023}, publisher = {Elsevier}, month = {}, }